A view into what’s really happening during gene editing for Precision CRISPR
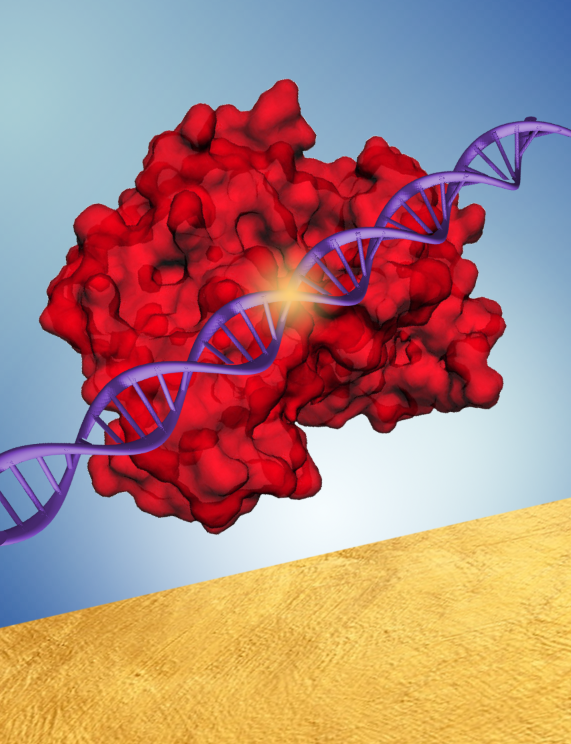
represents the bioplasmonic surface. Image: Somin Lee.
Gene editing is one of the hottest new techniques being explored by scientists and medical doctors alike, with the most typical applications including improved food production and treatment of serious diseases. Expected to reach a market size of $18.5M by 2028, the most popular method relies on the technique known as CRISPR (clustered regularly interspaced short palindromic repeats).
Prof. Somin Lee and her team have found a way to make CRISPR-style gene editing much more efficient and reliable thanks to a new technique that will allow doctors to view what’s happening in real time.
“This work paves the way for ‘Precision CRISPR’ to verify in real-time that every single cell is corrected,” said Lee.
CRISPR-Cas systems (ie, CRISPR-associated systems) are an extension of the CRISPR-Cas9 (CRISPR-associated protein 9) system, which was awarded The Nobel Prize in Chemistry in 2020. The technique was developed after understanding how bacteria come to destroy viruses. What happens is, after recognizing a destructive entity such as a virus, the bacteria mimic it enough to be able to attach itself to the virus. It then “cuts” the viral DNA apart, thereby destroying the defective gene.
Gene editing has already been used to improve the hardiness of plants, and the medical community has been experimenting with the technique to treat cancer, viral infections, and a variety of genetic diseases. It has already been successfully demonstrated in human cells with the goal of protecting the body against HIV, flu, and RSV.
Still, there is significant room for improvement. For example, a recent survey of “207 scientists using the technology states that on average it takes researchers 70+ weeks and 7+ failed experiments to obtain a successful CRISPR edit.”
In order to improve these numbers, scientists need to understand what’s happening throughout the entire process, which consists of conjugation (where genetic material is transferred from one bacterium to another through direct contact), target recognition, and final cleavage of the problematic genes, using real-time photonics.
This work paves the way for “Precision CRISPR” to verify in real-time that every single cell is corrected
Somin Lee
However, the norm is to simply wait and see if the process worked, and if not, keep trying. Relying on this trial and error method wastes valuable time, and makes it more difficult to intentionally impact the DNA in the way it was intended. It would be so much more helpful to be able to watch the process from beginning to end.
The most promising technique currently used to view the entire process is fluorescence imaging. Unfortunately, it yields only glimpses of what’s happening to the DNA because it’s also prone to photobleaching, which nullifies the fluorescence and sends the process back into darkness and invisibility.
As described in a recently published paper in Nanophotonics, Lee and her team overcame the negative impact of photobleaching by using single particle spectroscopy. As a result, they were able to view, for the first time, the complete CRISPR-Cas process on bioplasmonic surfaces.
And in the process, Lee’s team discovered why CRISPR-Cas efficacy tends to be quite low. The key problem lay in the fact that the target recognition and cleavage processes were surprisingly heterogeneous, which led to unsuccessful gene edits.
“Our technique can help the medical community use CRISPR-Cas systems more effectively in precision medicine and personalized disease therapeutics,” said Lee.
The paper, Dynamic observations of CRISPR-Cas target recognition and cleavage heterogeneities,” was authored by Zhijia Zhang (PHD student ECE), Haechan Jeong (MS ECE alumnus), Di Zu (PHD student, BME), Xintao Zhao (PHD student ECE), Pramith Senaratne (BS student, neuroscience), John Filbin (BS, Biochemistry), Brett Silber (BS, Neuroscience), Sarah Kang (BS student, neuroscience), Ann Gladstone (UG Biopsych, Cognit & Neurosci), Matthew Lau (BS, Biochemistry), Guangjie Cui (PHD student ECE), Young Geun Park (Asst Res Sci, ME) and Somin Eunice Lee.
Lee is also affiliated with the Departments of Biomedical Engineering, Molecular Science & Engineering, the Applied Physics program, and the Biointerfaces Institute at the University of Michigan.
 MENU
MENU 
